Expanding the SUSY analysis
Last updated on 2024-12-30 | Edit this page
Overview
Questions
- Can we use Snakemake and REANA in more complex examples?
Objectives
- Learn how to expand a SUSY analysis to handle more complex tasks using Snakemake and REANA.
Now that you’ve mastered the basics of creating simple Snakemake workflows and submitting them to REANA, let’s dive deeper into more complex analyses. This next example will illustrate how to expand our SUSY analysis to perform more sophisticated tasks.
A more real SUSY analysis
In real-world analysis workflows, processing often involves multiple
datasets. Typically, we identify these datasets using CERN’s Data Access
System (DAS) and then extract a list of files for further analysis.
These files are then processed individually before being merged for
subsequent steps. This workflow often involves additional CERN tools
like Rucio for dataset discovery
and manual steps, like using hadd to merge ROOT files.
However, Snakemake and REANA offer a powerful solution to automate and
streamline these tasks. Let’s explore how we can leverage their
capabilities to handle this complex scenario.
Remember
There isn’t a one-size-fits-all approach to creating workflows, just as there isn’t a single way to perform an analysis. The decisions made in this tutorial are primarily for illustrative purposes. While there may be more efficient or optimized ways to structure this workflow, we’ve chosen this approach to clearly demonstrate the core concepts involved.
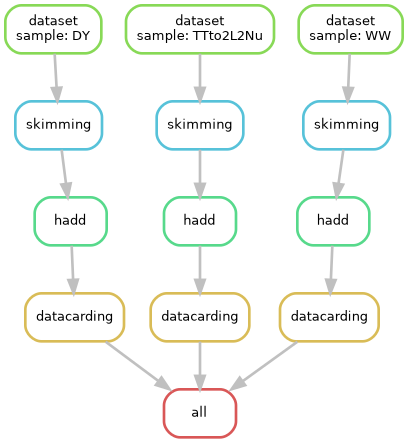
Let’s look at the Snakefile:
output_dir = "$REANA_WORKSPACE"
analysis_container = "docker://gitlab-registry.cern.ch/cms-analysis/analysisexamples/snakemake-reana-examples/cmsreana_susyexample:latest"
# Define the final target rule
rule all:
input:
expand("output/datacards/{sample}.root", sample=config["samples"])
# Rule for dataset
rule dataset:
output:
"output/dataset_{sample}.txt"
resources:
voms_proxy=True,
rucio=True
params:
sample = "{sample}"
container: "docker://docker.io/reanahub/reana-auth-rucio:1.1.1"
shell:
"""
source datasets.sh {params.sample} ### this is a workaround to get the dataset
mkdir -p {output_dir}/output/
rucio whoami
rucio list-file-replicas cms:$dataset --pfns --protocols root --rses "(tier=2|3)&(country=US|country=DE)" | head -n 5 > {output_dir}/output/dataset_{params.sample}.txt
head {output_dir}/output/dataset_{params.sample}.txt
"""
# Rule for skimming
rule skimming:
input:
"output/dataset_{sample}.txt"
output:
"output/skimming/{sample}/files.txt"
resources:
voms_proxy=True,
kerberos=True,
# compute_backend="htcondorcern"
params:
selection = '"(nMuon>0&&nTau>0&&HLT_IsoMu24)"',
N = 100,
sample = '{sample}'
container: analysis_container
shell:
"""
mkdir -p {output_dir}/output/skimming/{params.sample}
cd /code/CMSSW_13_0_10/src
source /cvmfs/cms.cern.ch/cmsset_default.sh
cmsenv
export X509_CERT_DIR=/cvmfs/grid.cern.ch/etc/grid-security/certificates
echo "Processing files listed in: {input}"
while IFS= read -r file; do
echo "Processing file: $file"
python3 PhysicsTools/NanoAODTools/scripts/nano_postproc.py \
{output_dir}/output/skimming/{params.sample}/ "$file" \
--bi /code/CMSSW_13_0_10/src/SUS_ex/Analysis/scripts/keep_in.txt \
--bo /code/CMSSW_13_0_10/src/SUS_ex/Analysis/scripts/keep_out.txt \
-c {params.selection} \
-I SUS_ex.Analysis.DiTau_analysis analysis_mutaumc \
-N {params.N} --prefetch
done < {output_dir}/{input}
find {output_dir}/output/skimming/{params.sample}/ -type f -name "*root" > {output_dir}/output/skimming/{params.sample}/files.txt
head {output_dir}/output/skimming/{params.sample}/files.txt
"""
# Rule for hadd
rule hadd:
input:
"output/skimming/{sample}/files.txt"
output:
"output/skimming/{sample}_Skim.root"
container: analysis_container
shell:
"""
mkdir -p {output_dir}/output/skimming
cd /code/CMSSW_13_0_10/src && \
source /cvmfs/cms.cern.ch/cmsset_default.sh && \
cmsenv && \
python3 PhysicsTools/NanoAODTools/scripts/haddnano.py \
{output_dir}/output/skimming/{wildcards.sample}_Skim.root \
$(cat {output_dir}/{input})
"""
# Rule for datacarding
rule datacarding:
input:
"output/skimming/{sample}_Skim.root"
output:
"output/datacards/{sample}.root"
params:
year = "2022postEE",
sample = '{sample}'
# sample = lambda wildcards: wildcards.sample
container: analysis_container
shell:
"""
mkdir -p {output_dir}/output/datacards
cd /code/CMSSW_13_0_10/src/SUS_ex/Analysis2 && \
source /cvmfs/cms.cern.ch/cmsset_default.sh && \
cmsenv && \
./FinalSelection_mutau.exe {params.year} {output_dir}/{input} {output_dir}/{output} {params.sample} {params.sample}
"""
Before running this workflow, let’s discuss a few new features introduced in this example:
- Rule-Specific Containers: One of the advantages of containerization is the ability to define specific software environments for each rule. This allows you to use different tools and configurations for different parts of your workflow.
- Consolidated Output Folder: While not strictly necessary, organizing all outputs into a single folder can simplify your workflow and subsequent analysis. We recommend this approach for better organization and easier management.
- Diverse Resource Requirements: To access CMS datasets, Rucio, or other resources, you often need to configure specific credentials and settings. REANA seamlessly integrates with these resources, allowing you to specify the necessary resources for each rule.
REANA resources
Securing Your Credentials with REANA Secrets
To access resources like CMS datasets or Rucio, you’ll need to
provide your credentials, such as VOMS proxies. To ensure the security
of this sensitive information, REANA uses secrets. These
secrets are encrypted and only accessible to your workflow, protecting
your credentials from unauthorized access.
For detailed instructions on how to configure secrets in REANA, please refer to the official REANA documentation:
- VOMS-PROXY: instructions
- kerberos: instructions
- Rucio: instructions
Users can verify which secrets REANA know by running in the terminal:
IMPORTANT INFORMATION ABOUT VOMS-PROXY
The REANA documentation outlines two methods for configuring VOMS-PROXY credentials:
- Automatic Generation: (Recommended approach) Leverages your user certificate and key to automatically generate the VOMS-PROXY when needed. Ensures that your credentials are always up-to-date and valid.
- Manual Upload: Requires you to upload your VOMS-PROXY file to REANA. Less convenient as you’ll need to re-upload the file whenever it expires.
Important: Do not use both methods simultaneously. REANA prioritizes the manually uploaded VOMS-PROXY file, so if it’s expired, you won’t be able to access remote files via XRootD.
We strongly recommend using the automatic generation method for a seamless and secure workflow.
Computer backends
By default, REANA utilizes Kubernetes as its computing backend. This allows you to easily scale your workflows and allocate resources as needed. For instance, to increase the memory allocated to a specific rule, you can specify it in the resources section:
rule skimming:
...
resources:
compute_backend="kubernetes",
kubernetes_memory_limit="8Gi",
...Note that Kubernetes imposes a maximum memory limit of
9.5Gi per container.
For more demanding workloads or specific resource requirements, REANA also supports integration with HTCondor and Slurm. These backends can provide access to larger computing resources and more flexible scheduling options, but their usage is outside the scope of this tutorial. Please refer to the official documentation for more information.
Storage backends
Each REANA user is allocated 300GB of storage space on the platform. (This is visible in under your profile in the web interface). While this is typically sufficient for most workflows, you can leverage REANA’s integration with your personal EOS storage area for larger datasets. To enable this integration, ensure that your Kerberos credentials are configured correctly in your REANA profile.
In this case, the workflow should work perfectly.
More REANA examples
The REANA team has a compilation of examples in their github area. It is well recommended to visit them for more inspiration.
In addition, users can seek for support and help in the following channels:
- REANA official mattemost channel
- REANA forum
- CMS-REANA mattermost channel
- Explore the REANA team’s compilation of examples on their GitHub for more inspiration and advanced use cases.
- Utilize the REANA support channels, such as the official Mattermost channel and forum, for assistance and community support.
